About Me
I am a principal investigator at Aithyra in Vienna, Austria. Aithyra is a new research institute at the intersection of machine learning and life sciences led by Michael Bronstein and funded by the Boehringer Ingelheim Foundation. If you’re interested in PhD, Postdoc, or Visiting researcher positions please feel free to reach out via email! PhD applications for fall 2026 will open soon.
Previously, I was briefly an assistant professor at Duke University. Before that I did my postdoc with Yoshua Bengio working on efficient machine learning algorithms with applications to cell and molecular biology at Mila in Montreal. I completed my PhD in the computer science department at Yale University in 2021 where I was advised by Smita Krishnaswamy. My dissertation can be found here. My research interests are in generative modeling, deep learning, and optimal transport. I’m working on applying ideas from generative modeling, causal discovery, optimal transport, and graph signal processing to understand how cells develop and respond to changing conditions. I’m also interested in generative models for protein design and cofounded Dreamfold to work on these problems.
I grew up in Seattle and graduated from Tufts University in 2017 with a BS and MS in computer science. Outside of work, I love sailing and running. I am the 2019 junior North American champion in the 505 class, and I recently ran my first 50 mile race the Vermont 50!
Recent News
- Joined Aithyra as a principal investigator. If you’re interested in PhD, Postdoc, or Visiting positions please feel free to reach out via email!
- Congrats to Danyal Rehman and collaborators for winning the Best Paper Award at the Gen Bio workshop at ICML 2025 for our work on forward only regression training of normalizing flows FORT.
- Join us at the second edition of the Frontiers in Probabilistic Inference: Sampling meets Learning workshop at NeurIPS 2025 in San Diego.
- Started as an assistant professor at Duke University.
- Two papers accepted to ICML 2025. Checkout our work on Feynman-Kac steering of diffusion models (spotlight) and Scaling Boltzmann Generators as well as newer workshop papers such as FORT, PITA and other works coming soon.
- Congrats to Fred Zhangzhi Peng and collaborators for winning an outstanding paper award at the DELTA workshop at ICLR 2025 for our work on improved sampling from masked diffusion models in P2.
- Four papers accepted to ICLR 2025. Checkout our work on steering masked and continuous diffusion and our work on flow matching in cells with greater realism in CFGen and transferrability in Meta FM.
- Join us at our workshop Frontiers in Probabilistic Inference: Sampling meets Learning at ICLR 2025 in Singapore.
Older News (2024)
- Presenting a tutorial on Geometric Generative Models with Heli Ben-Hamu and Joey Bose at the LoG Conference 2024.
- Visiting the group of Michael Bronstein at Oxford winter 2024.
- Three papers accepted to NeurIPS 2024! Awesome work led by Xi (Nicole) Zhang, Yuan Pu, Guillaume Huguet, James Vuckovic, and Kacper Kapuśniak.
- Generative Modeling
- Flow Models
- Optimal Transport
- Protein Design
- Single Cell Dynamics
Postdoc
Mila & University of Montreal
PhD in Computer Science, 2021
Yale University
MPhil in Computer Science, 2020
Yale University
MS in Computer Science, 2017
Tufts University
BS in Computer Science, 2017
Tufts University
Featured Publications
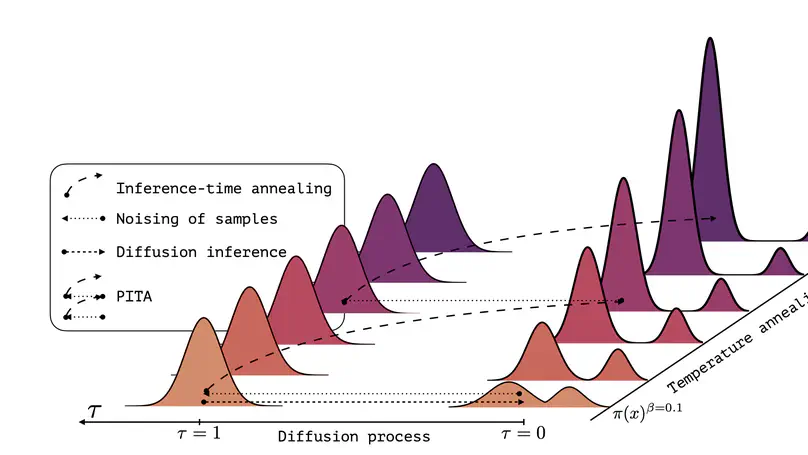
Sampling efficiently from a target unnormalized probability density remains a core challenge, with relevance across countless high-impact scientific applications. A promising approach towards this challenge is the design of amortized samplers that borrow key ideas, such as probability path design, from state-of-the-art generative diffusion models. However, all existing diffusion-based samplers remain unable to draw samples from distributions at the scale of even simple molecular systems. In this paper, we propose Progressive Inference-Time Annealing (PITA), a novel framework to learn diffusion-based samplers that combines two complementary interpolation techniques: I.) Annealing of the Boltzmann distribution and II.) Diffusion smoothing. PITA trains a sequence of diffusion models from high to low temperatures by sequentially training each model at progressively higher temperatures, leveraging engineered easy access to samples of the temperature-annealed target density. In the subsequent step, PITA enables simulating the trained diffusion model to procure training samples at a lower temperature for the next diffusion model through inference-time annealing using a novel Feynman-Kac PDE combined with Sequential Monte Carlo. Empirically, PITA enables, for the first time, equilibrium sampling of N-body particle systems, Alanine Dipeptide, and tripeptides in Cartesian coordinates with dramatically lower energy function evaluations. Code available at: https://github.com/taraak/pita
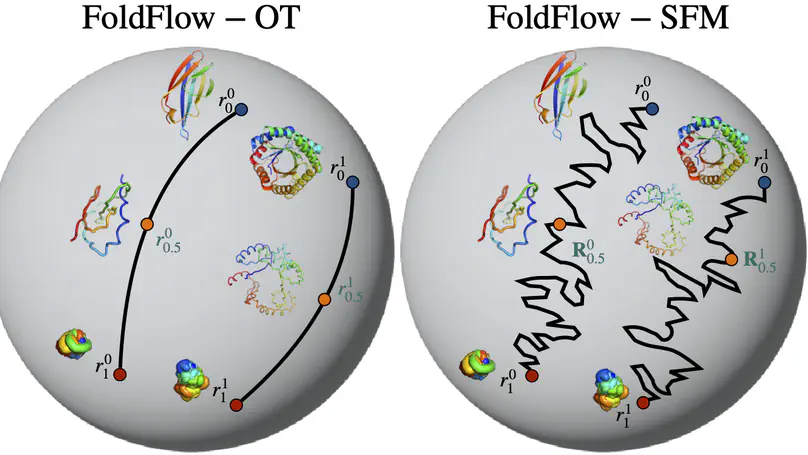
We present FoldFlow a novel flow matching model for protein design. We present theory and practical tricks for flow models over SE(3)^N. Empirically, we validate these models on protein backbone generation of up to 300 amino acids leading to high-quality designable, diverse, and novel samples.
Publications
Also presented at Frontiers4LCD Workshop @ ICML 2023.
PDF Cite DOI code reproducible code Tutorial Cancer Research Spotlight Yale News News-Medical
Also presented at Frontiers4LCD Workshop @ NeurIPS 2022.
Also presented at Frontiers4LCD Workshop @ ICML 2023.
Also presented at ML4M Workshop @ NeurIPS 2020.
Also presented at LMRL Workshop @ NeurIPS 2020.
PDF Cite Code Poster Slides Video Arxiv ICML Page Tweetorial Workshop Poster
Also at GRLB Workshop @ ICML 2020.
Best Student Paper Award.
PDF Cite Code Slides Video Yale Daily Article Best Paper Award
Also at LMRL Workshop @ NeurIPS 2019.
PDF Cite Code Project Slides Video Arxiv ICML Page Introductory Talk
Also presented at RLGM Workshop @ ICLR 2019.